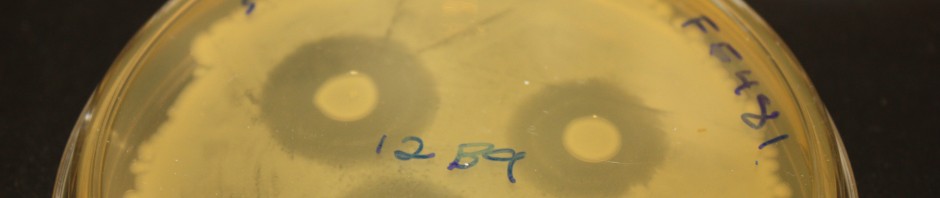Basalla J, Harris R, Burgess E, Zeedyke N, and Wildschutte H. Expanding Tiny Earth to genomics: a bioinformatic approach for an undergraduate class to characterize antagonistic strains. FEMS Microbiol Letters. Jan 2020; 367(1):18fnaa. doi: 10.1093/femsle/fnaa018
Karunarathna MHJS, Bailey KM, Ash BL, Matson PG, Wildschutte H, Davis TW, Midden WR, Ostrowski AD. Nutrient Capture from Aqueous Waste and Photocontrolled Fertilizer Delivery to Tomato Plants Using Fe(III)-Polysaccharide Hydrogels. ACS Omega. Sep 2020;5(36):23009-23020. doi: 10.1021/acsomega.0c02694.
Mechan-Llontop ME, Tian L, Bernal-Galeano V, Wildschutte H, Marine SC, Yoder KS, and Vinatzer BA. Exploring rain as source of biological control agents for fire blight on apple. Frontiers Microbiol. Feb 2020; 11:119. doi.org/10.3389/fmicb.2020.00199.
Basalla B, Chatterjee P, Burgess E, Khan M, Verbrugge E, Wiegmann DD, LiPuma JJ, and Wildschutte H. Loci encoding compounds potentially active against drug resistant pathogens amidst a decreasing pool of novel antibiotics. Appl Environ Microbiol. Dec 2019; 85(2):e01438-19. doi: 10.1128/AEM.01438-19
Wagner A, Norris S, Chatterjee P, Morris P, and Wildschutte H. Aquatic pseudomonads inhibit oomycete plant pathogens of Glycine max. Frontiers Microbiol. May 2018; 29:9:1007. doi.org/10.3389/fmicb.2018.01007
Burks DJ, Norris S, Kauffman K, Joy A, Arevalo P, Azad RK, and Wildschutte H. Environmental vibrios inhibit human pathogenic Vibrio cholerae and Vibrio parahaemolyticus. MicrobiologyOpen. Aug 2017; e504. doi: 10.1002/mbo3.504.
Davis E, Sloan T, Tripathi A, Aurelius K, Barbour A, Bodey E, Clark B, Dennis C, Drown R, Fleming M, Humbert A, Kerns T, Lingro K, McMillin M, Meyer A, Pope B, Stalevicz A, Steffen B, Steindl A, Williams C, Wimberley C, Zenas R, Butela K, and Wildschutte H. Antibiotic discovery though The Small World Initiative: a molecular strategy to identify biosynthetic gene clusters involved in antagonistic activity. MicrobiologyOpen. Jan 2017; 00:1-9. doi: 10.1002/mbo3.435
Chatterjee P, Davis E, Yu F, James S, Wiegmann DD, Sherman DH, McKay RM, LiPuma JJ, and Wildschutte H. Environmental pseudomonads inhibit cystic fibrosis patient-derived Pseudomonas aeruginosa. Appl Environ Microbiol. Jan 2017; 83(2):e02701-16. doi: 10.1128/AEM.02701-16
D’souza NA, Kawarasaki Y, Lee RE, Gantz JD, Beall BFN, Shtarkman YM, Koçer ZA, Rogers SO, Wildschutte H, Bullerjahn GS, McKay RML. Microbes promote ice formation in large lakes. ISME J. Aug 2013; 7(8):1632-1640. doi: 10.1038/ismej.2013.49
*Cordero OX, *Wildschutte H, *Kirkup B, Proehl S, Hussain F, Ngo L, Le Roux F, Mincer T, and Polz MF. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. Sept 2012; 337:1228-1231.
doi: 10.1126/science.1219385. *contributed equally.
Preheim SP, Boucher Y, Wildschutte H, David L, Veneziano D, Alm E, Polz, MF. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol. May 2011; 13(1):265-75. doi: 10.1111/j.1462-2920.
Wildschutte H, Preheim SP, Hernandez Y, and Polz MF. O-Antigen Diversity and Lateral Gene Transfer of the wbe Region Among Vibrio splendidus Isolates. Environ Microbiol. Nov 2010; 12(11):2977-87. doi: 10.1111/j.1462-2920.
Dedrick RM, Wildschutte H, McCormick JR. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J Bacteriol. 2009 Jan;191(1):320-32. doi: 10.1128/JB.00858-08
Wildschutte H and Lawrence JG. Differential Salmonella survival against communities of intestinal amoebae. Microbiology 2007 Jun;153(Pt 6):1781-9. doi: 10.1099/mic.0.2006/003616-0
Wildschutte H, Wolfe DM, Tamewitz A, and Lawrence JG. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc Natl Acad Sci USA. 2004 Jul 20; 101(29):10644-9. doi: 10.1073/pnas.0404028101
Guillot J, Demanche C, Norris K, Wildschutte H, Wanert F, Berthelemy M, Tataine S, Dei-Cas E, Chermette R. Phylogenetic relationships among Pneumocystis from Asian macaques inferred from mitochondrial rRNA sequences. Mol Phylogenet Evol. 2004 Jun;31(3):988-96. doi: 10.1016/j.ympev.2003.10.022
Norris KA, Wildschutte H, Franko J, Board KF. Genetic variation at the mitochondrial large-subunit rRNA locus of Pneumocystis isolates from simian immunodeficiency virus-infected Rhesus macaques. Clin Diagn Lab Immunol. 2003 Nov;10(6):1037-42. PMCID: PMC262427